Gene Ontologies
This web page was produced as an assignment for Genetics 677, an undergraduate course at University of Wisconsin: Madison.
In order to find GO terms that were used to discribe the gene DCDC2 I attempt to use 5 databses to see if I got the same or different results.
AMiGO:
When using Amigo, four entries were found upon searching for DCDC2: two entries were cdc2 and other two entries were DCDC2, one was in Homo sapiens and other one was in Mus musculus.
DCDC2: Homo sapiens
Biological process: Cellular Defense Response: A defense response that is mediated by cells.
Accession Number:GO:0006968
Number of gene products: 91
Neuron migration: The characteristic movement of immature neurons from germinal zones to specific positions where they will reside as they mature.
Accession number: GO:0001764.
Number of gene products 154
DCDC2a: Mus musculus. It has only the biological process, cellular component and molecular function listed.
GOA:
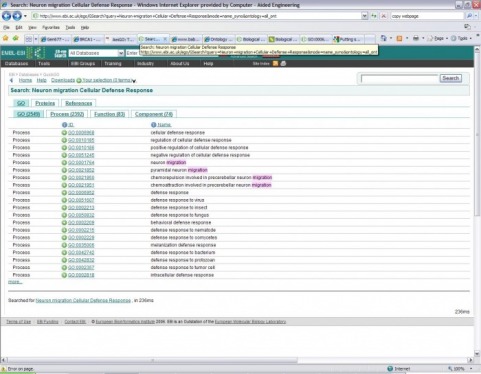
After using amigo, I used GOA and used the search terms: Neuron migration Cellular Defense Response. This is what appeared:
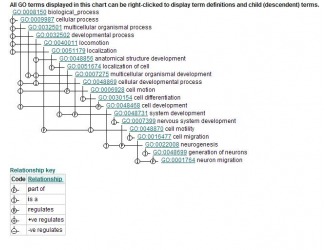
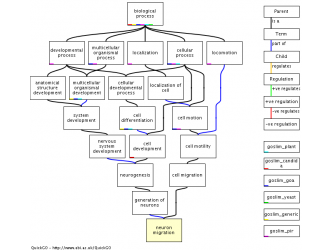
Then I clicked on the neuron migration GO: 0001764. This displayed the page known as GO:0001764 neuron migration. This page had tabs that were labeled as: term information, ancestor chart, ancestor terms, child terms, protein annotations, and statistics. The ancestor chart, and terms had very good images.
Figure 2. Ancestor table from GOA Figure 3. Acesntor Chart from GOA
GOALL:
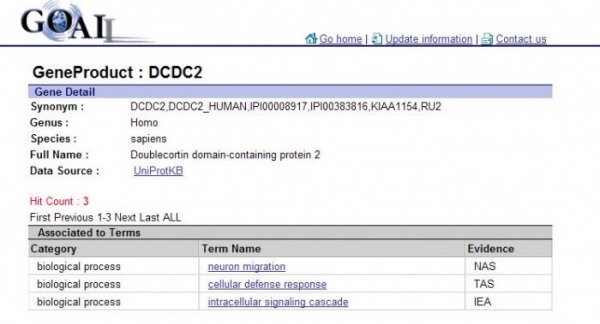
This database allows one to search GO ID, terms, and Gene. I used the gene search DCDC2 and got a very nice presentation in a table. A GO term that did not appear in AMiGO that was found with this database was intracellular signaling cascade.
Figure 4. Webpage picutre of a search in GOALL using the gene nameDCDC2
Gramene:
This database is only useful if you know your GO terms or the plant homolog of you gene name. This database does have one of the better tutorials, however it is strictly plants and grass.
QuickGO:
It allows you to do search using gene name, better of with the protein name better with this, and the Go terms. The search terms I used in a single search were Cellular Defense Response, GO:0006968, Neuron migration, GO: 0001764. I also used the search terms DCDC2 and RU2.
Analysis
I found all the databases useful except the Gramene database. I did not find this one useful because I do not know the homolog to the DCDC2 gene or its protein that exists in plants. Otherwise this database seems very manageable. However, I did a GO terms search instead, and this database seems very well laid out and useful to people who know more about plant genes than me.
I found the AmiGo database was the place to start when searching just based on your gene interest. It gives a very good list of all the GO terms that are associated with your gene of interest and you are able to get the definitions right at your fingertips. I would like to make a note that my gene of interest (DCDC2) has not had much research done on it besides its associations with dyslexia, so its number of GO terms and detail may be a bit more limited compared to more common and longer researched genes. It also has a limited number of GO terms and other associations.
All of the 5 database listed above can be used to derive many more GO association. The databases AmiGo, GOA, GOALL, and QuickGo allow one to find GO terms by searching for the gene of interest. I found that AmiGo, GOA, GOALL were easier to use to find GO terms when using you gene interest compared to QuickGO. QuickGo required one to know a lot about the gene, such as synonyms for the gene or protein. I got different results when I searched for DCDC2 in QuickGO compared to RU2 in QuickGO. When I used DCDC2 I got the zebra fish doublecortin protein and when I used RU2 I got the Homo sapiens DCDC2 gene. All the databases had a different ways of visually presenting the data and terminology. I personally liked the GOALL presentation of the information since in my opinion it is laid out in a visually pleasing and simple manner. I thought the QuickGO was little more complicated since I did not get the Homo sapiens result I needed when searching DCDC2, but I got it when I searched for RU2 instead.
However, I liked the fact that GOA had excellent graphs and tables that laid out the terms associations. I found that all the databases found the basically same three GO terms for my gene and covered all the relevant categories. Thus, a person can use the only one database when searching for Go terms and associations if they want depending on their work, knowledge, what they are specifically looking for, and what database they find more manageable and visually pleasing.
References of databases:
AmiGo
GOA
GOALL
Gramene
QuickGo
Images:
(Figure1) http://www.ebi.ac.uk/ego/GSearch?query=Neuron+migration+Cellular+Defense+Response&mode=name_syno&ontology=all_ont
(Figure2) http://www.ebi.ac.uk/ego/GTerm?id=GO:0001764
(Figure3) http://www.ebi.ac.uk/ego/GTerm?id=GO:0001764
(Figrue4 ) http://www.shigen.nig.ac.jp/ontology/geneDetailAndTermListAction.do?geneProductId=97661
Megan Holler
[email protected]
Last Updated: 2/12/09